flika: image processing for biologists¶
flika makes it easy to write analyze images and movies of biological data. It is quick to get started, and is easy to extend by installing a wide variety of plugins or writing your own.
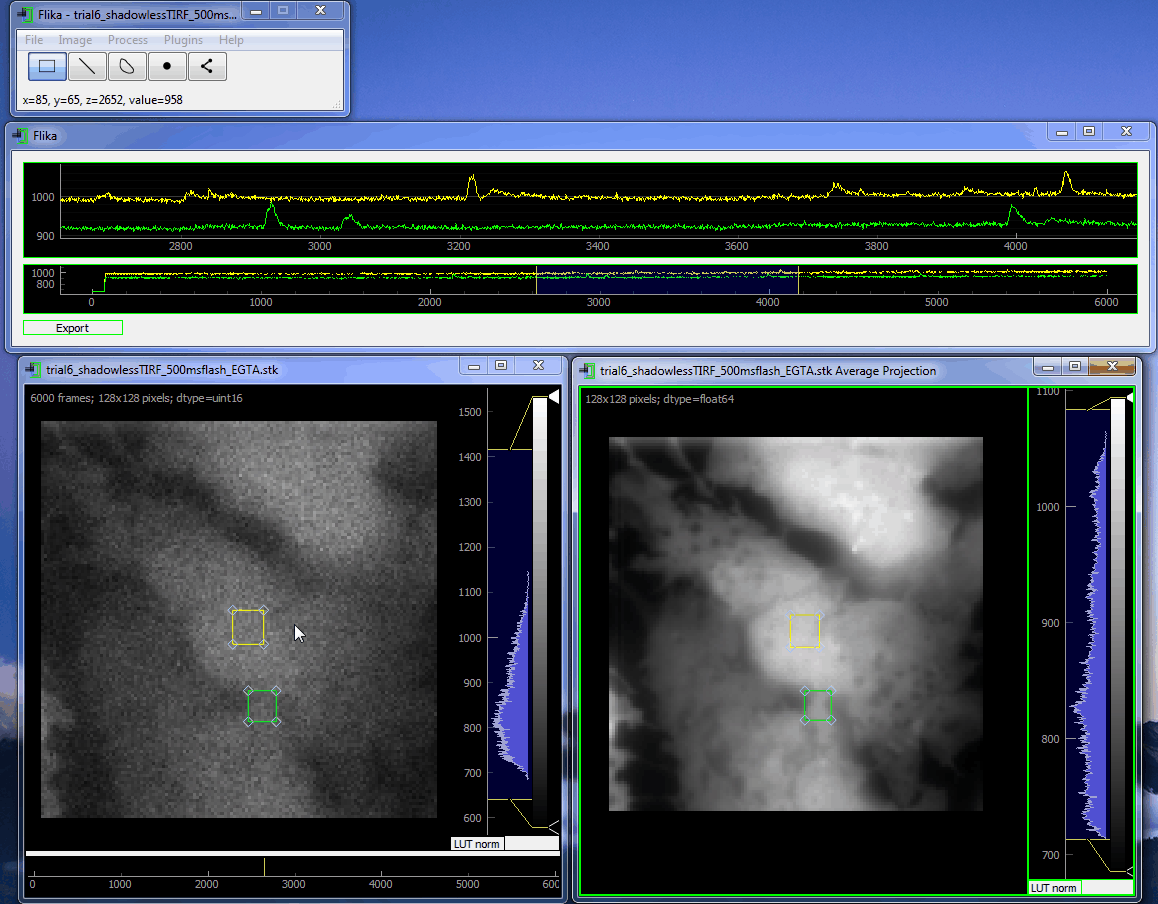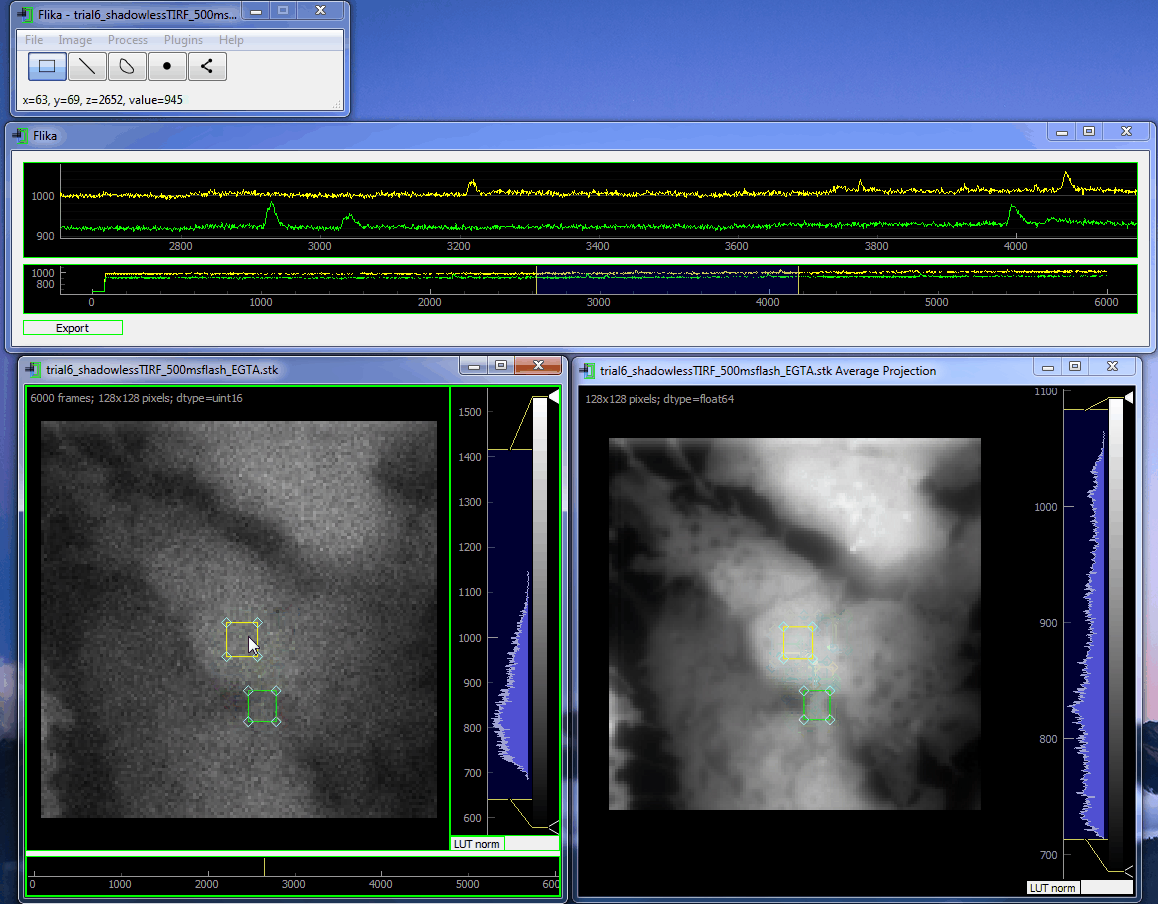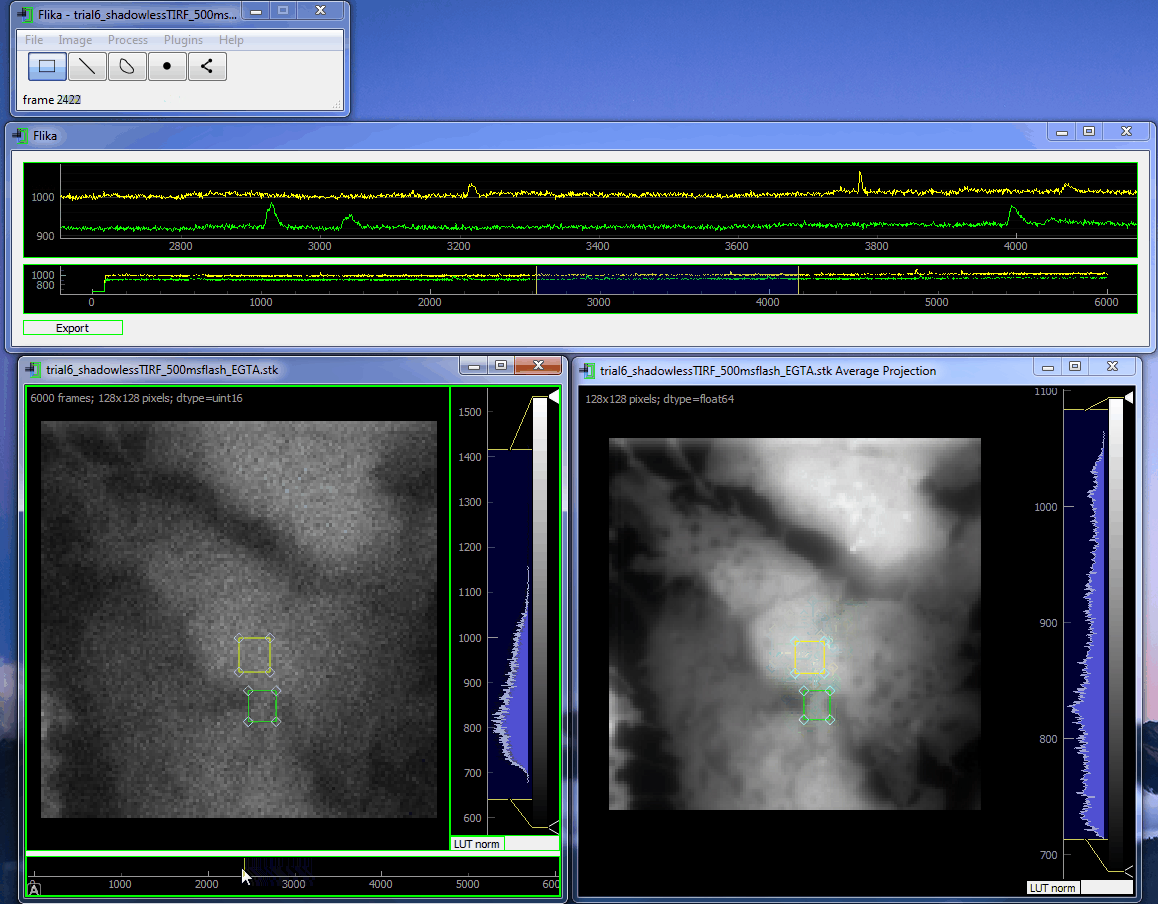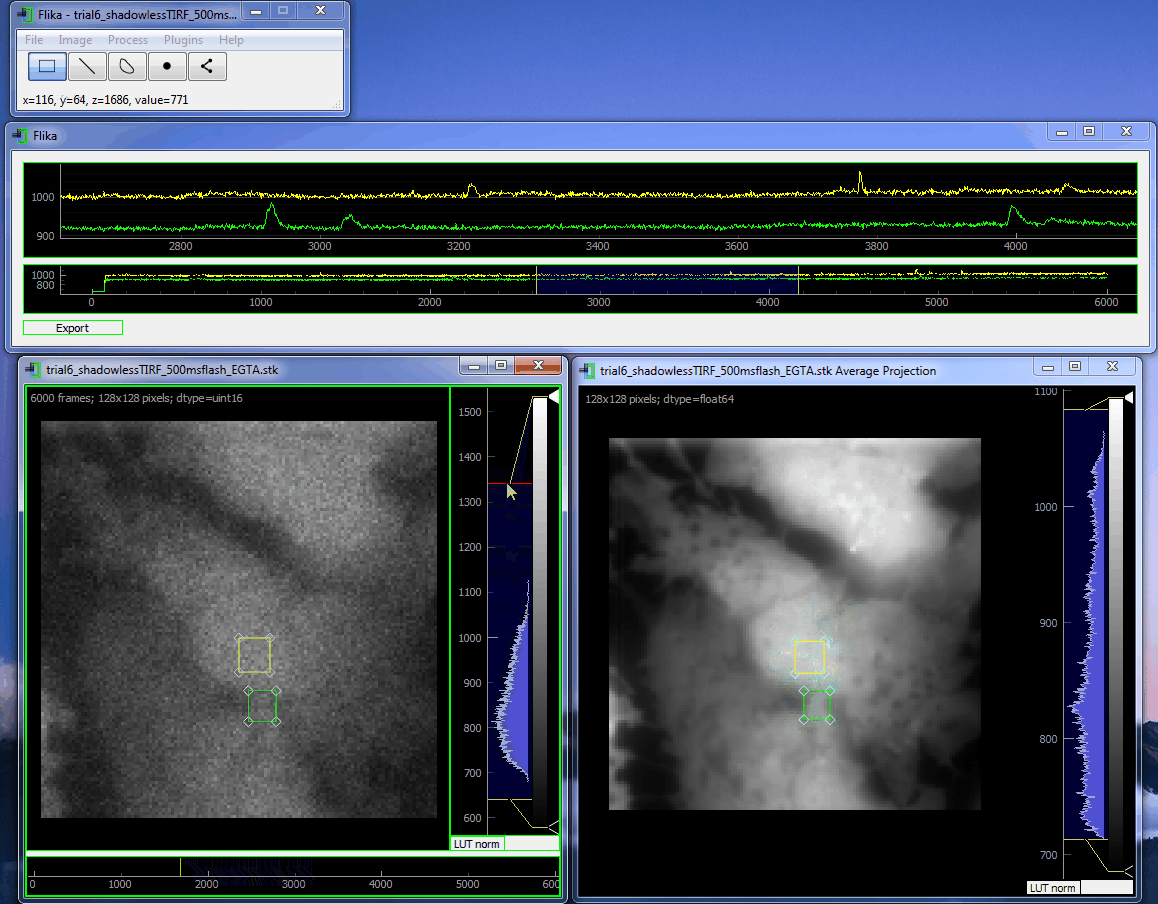
Install¶
Please see Installation for instructions. flika runs on Windows, OSX, and Linux.
Features¶
- Supports numerous types of image filetypes: .tif, .nd2, .stk, and more.
- Record the steps of your analysis in a script to automate a series of flika commands.
- Python 3
- Rich plugin architecture, with many external plugins and a growing community.
- Open source under the MIT license.
Bugs/Requests¶
Please use the GitHub issue tracker to submit bugs or request features.
Citations¶
flika was created to automatically identify and automate the analysis of local Ca 2+ signals but has grown into a general analysis tool for studying biological images. When using flika, please cite:
Ellefsen, K., Settle, B., Parker, I. & Smith, I. An algorithm for automated detection, localization and measurement of local calcium signals from camera-based imaging. Cell Calcium. 56:147-156, 2014
Supported through National Institutes of Health grants GM 100201 to Ian Smith, and GM 048071 and 065830 to Ian Parker.
Authors¶
So far, code has been contributed by Kyle Ellefsen , Brett Settle and Kevin Tarhan. If you would like to add your name to the list, see our guide for contributing.
License¶
Distributed under the terms of the MIT license, flika is free and open source software.
